





Young Lab
Compensatory mechanisms in eye development and disease.
Young Lab
Compensatory mechanisms in eye development and disease.
About Us
Our research aims to identify and understand the function of genes required for eye formation and study the cellular and molecular compensatory mechanisms that enable phenotypic robustness in eye development.
We use genetic tools/screens in zebrafish to identify the genes that participate in eye formation; and embryology, cell biology, and molecular techniques to understand their function.
We are based at the Institute of Ophthalmology, a world leading centre for eye research.
Young Lab News

Our Research
Our Research
OUR RESEARCH
The eye is an essential organ for animal viability, and as such its appropriate development and function is fundamental for life. For this reason, biological systems have evolved developing mechanisms that enable tissues/organs to compensate for the effect of mutations in genes, preventing their impact on eye development.
Compensatory mechanisms that enable the systems’ robustness can mask the effect of mutations in many genes, showing no overt morphological phenotype and limiting our ability to understand their function. Therefore, relating a phenotype or pathology to a gene represents a major challenge in developmental genetics, and suggests that many genes involved in embryo development or disease remain to be identified.
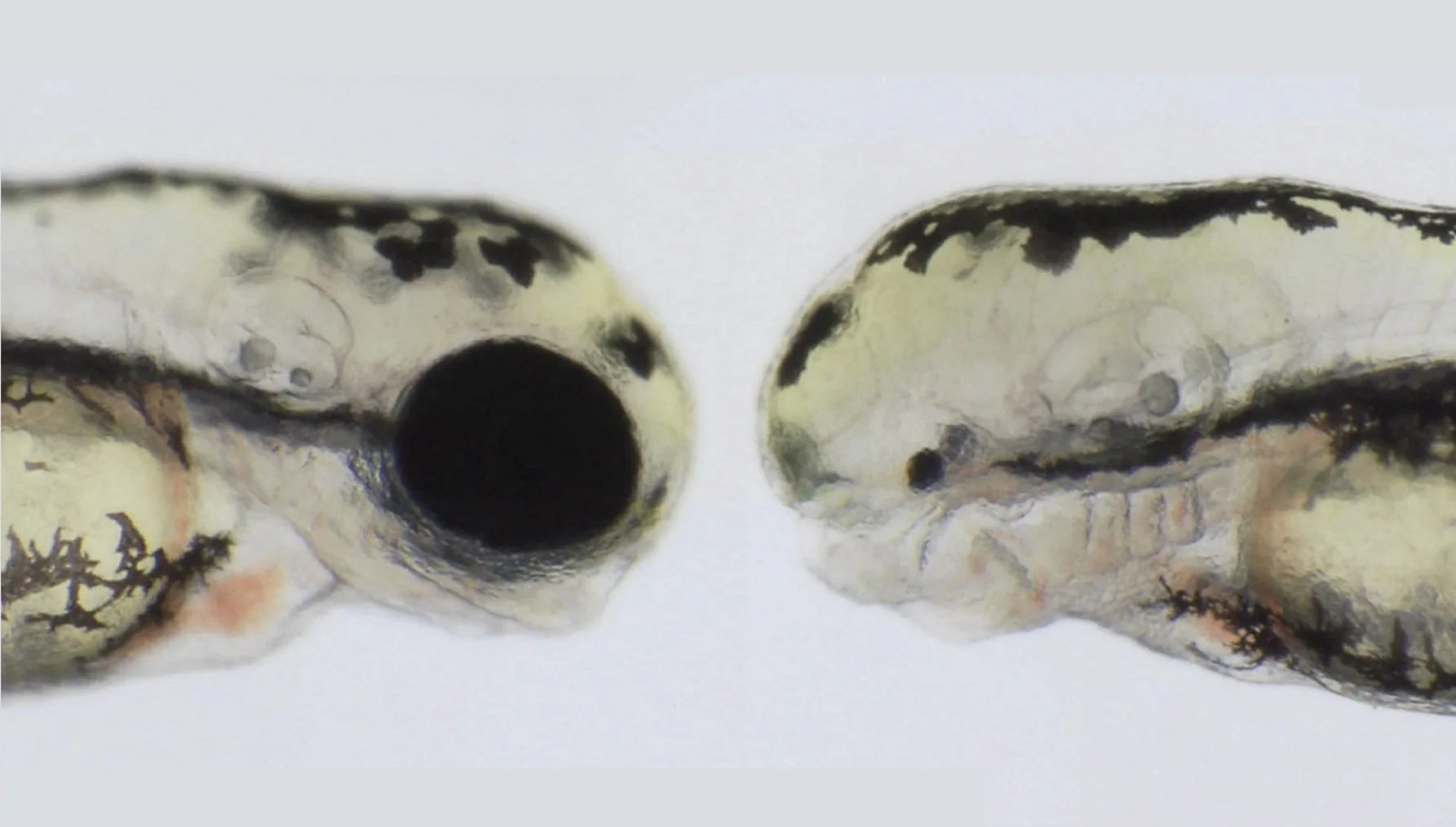
For example, due to compensatory mechanisms, only ~30% of all the genes that have been linked to anophthalmia (A, no eyes) and microphthalmia (M, small underdeveloped eyes) show a clear eye phenotype when knocked down in animal models. This limits the use of animal models to validate new A/M candidate genes identified in patients, or studying the cellular and molecular mechanisms behind these congenital malformations.
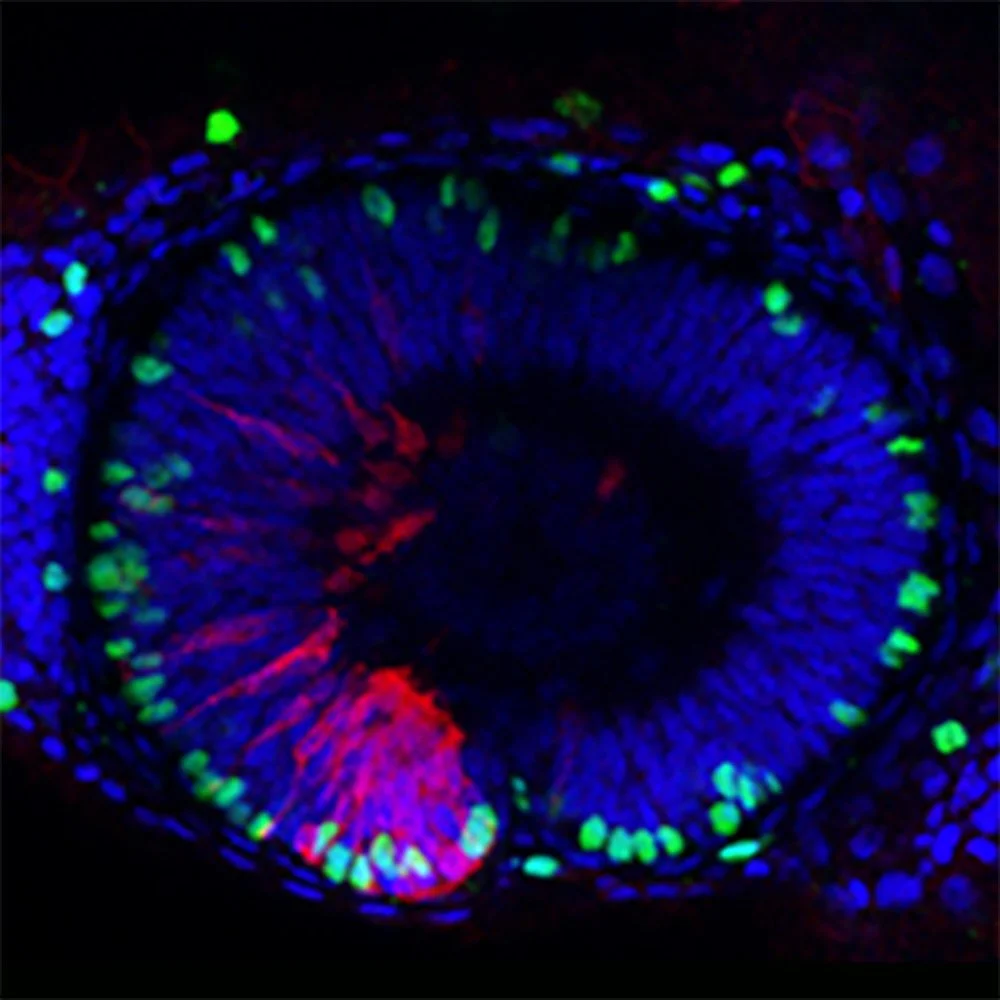
Analysis of mechanisms that enable growth compensation in eye development
Eye growth compensation in zebrafish (Young et al., 2019), allows the eyes to reach their expected size even when they start developing from a primordium with half the number of cells, and can mask the effect of mutations that lead to a smaller eye primordium.
Our lab studies the cellular and molecular mechanisms that modulate the balance of proliferation and differentiation of retinal progenitors that enable eye growth compensation. We also study new cellular and molecular functions of genes involved in eye formation, in which mutations give rise to congenital eye globe defects in humans.
Molecular diagnosis of anophthalmic and microphthalmic patients
Anophthalmia and microphthalmia has been linked to mutations in approximately 70 genes. The genetic diagnosis of these patients allows timely genetic counselling to improve their quality of life. Our lab is part of the Research Network with access to the Genomics England 100K Genomes Project database, which harbours the whole genome sequence of thousands of rare disease patients with no successful molecular diagnosis. We are currently using new pipelines to analyse the genome of A/M patients to identify the causal mutations aiming to identify new genes linked to this disease. We are using zebrafish to validate and address the function of the candidate genes we identify in A/M patients.

Outreach
Outreach
Outreach
This is a 15-minute cut of a longer documentary, including our lab’s research (min 11.43), showcasing some of the beauty and translatability of developmental biology research being undertaken currently throughout labs in the UK. Alice Roberts talks us through the wonder of developmental biology, and Courtney Lancaster (and her twin sister) tour various labs and interview PIs working with several model organisms to find out what drives various elements of embryo development, and how their studies may one day impact the clinic.

People
People
Current Lab Members
Collaborators
Gavin Arno (Greenwood Genetic Center, US)
Brian Brooks (NIE, US)
Genomics England (UK)
Lev Parsov (U. Michigan, US)
Florencia Cavodeassi (St George's University, UK)
Juan Ramon Martinez-Morales (CABD, Seville, Spain)
Previous Lab members
Cesar Valencia - Marine Biology undergraduate project, UNAB, Chile (2025)
Haowen Xue - UCL Biomedical Sciences, Summer Project.(2025)
Lucas Spink - BSc Biomedical Sciences undergraduate project, UCL (2024-25)
Lean Abahreh - MSc project UCL - Genetics of Human Disease (2024-25)
Cristian Aedo - Biotechnology undergraduate project, Universidad Santo Tomas, Chile (2023-24)
María Elisa Cuevas - Biotechnology Engineering undergraduate project, Universidad Católica, Chile (2023-24)
Pablo Paillalí - Research Assistant (2023-24)
Peilin Chen - BSc Biomedical Sciences undergraduate project, UCL (2023-24)
Dr Manuela Lahne - MRC project Postdoc (2023-24)
Robin Hayes - MSc project UCL - Genetics of Human Disease (2022-23)
Karthika Jeganathan –MSc project UCL - Genetics of Human Disease (2021-22)
Dr Montserrat Olivares – Postdoc, EMBO short-term fellow (2021)
Aphena Huang – MSc project UCL - Genetics of Human Disease (2020-21)

Publications
Publications
Selected Publications
Mara I. Maftei, Lucas G.N. Spink, Oriol Gracia Carmona, Simona Mikula Mrstakova, Lean Abahreh, Robin Hayes, Aitor Banon, Maria Elisa Cuevas, Katherine Cid, Raul Araya-Secchi, Franca Fraternali, Jing Yu, Gavin Arno, Rodrigo M. Young (2025)
Refining the genetic landscape of anophthalmia and microphthalmia: a comprehensive framework with deep learning and updated gene panels
medRxiv 2025.08.26.25334245; doi: https://doi.org/10.1101/2025.08.26.25334245
Neelathi UM, Ullah E, George A, Maftei MI, Boobalan E, Sanchez-Mendoza D, Adams C, McGaughey D, Sergeev YV, Ai Rawi R, Naik A, Bender C, Maumenee IH, Michaelides M, Tan TG, Lin S, Villasmil R, Blain D, Hufnagel RB, Arno G, Young RM, Guan B, Brooks BP. (2025)
Variants in NR6A1 cause a novel oculo vertebral renal syndrome.
Nat Commun. 2025 Jul 3;16(1):6111. doi: 10.1038/s41467-025-60574-y.
Monfries C. Carter, C, Ataliotis P, Bseisu A, Shaikh M, Hernández-Bejarano M, Fourteia M, Maftei MI, Young RM, Wilson SW, Gestri G and Cavodeassi F. (2025)
fzd5 zebrafish mutants behave as a sensitised genetic background prone to develop microphthalmia and coloboma.
Dis Model Mech. 18 (6): dmm052284. doi.org/10.1242/dmm.052284.
Powell, G.T., Faro, A., Zhao, Y., Stickney, H., Novellasdemunt, L., Henriques, P., Gestri, G., Redhouse White, E., Ren., J., Lu, W., Young, R.M., Hawkins, T.A., Cavodeassi, F., Schwarz, Q., Dreosti, E., Raible, D.W., Li, V.S.W., Wright, J.W., Jones, Y. and Wilson, S.W. (2024)
Cachd1 interacts with Wnt receptors and regulates neuronal asymmetry in the zebrafish brain.
Science. 384(6695):573-579. doi: 10.1126/science.ade6970.
Young, R.M,* Cavodeassi, F., Hawkins, T.A., Stickney, H.L., Schwarz, Q.P., Lawrence, L.M., Wierzbicki, C., Cheng, B.Y.L., Luo, J., Ambrosio, E.M., Klosner, A., Sealy, I.M., Rowell, J., Trivedi, C.A., Bianco, I.H., Allende, M.L., Busch-Nentwich, E.M., Gestri, G. and Wilson, S.W. (2019) Compensatory mechanisms render Tcf7l1a dispensable for eye formation despite its cell-autonomous requirement in eye field specification.
eLife doi:10.7554/eLife.40093. *Co-corresponding author.
Young, R.M,* Ewan, K.B., Ferrer, V.P., Allende, M.L., Godovac-Zimmermann, J., Dale, T.C., Wilson, S.W. (2019)
Developmentally regulated tcf7l2 alternative splice variants mediate transcriptional repressor functions during eye formation.
eLife doi:10.7554/eLife.51447. *Co-corresponding author.
Holt, R.J.*, Young, R.M.*, Crespo, B., Ceroni, F., Curry, C.J., et al. (2019)
De Novo Missense Variants in FBXW11 Cause Diverse Developmental Phenotypes Including Brain, Eye, and Digit Anomalies.
Am J Hum Genet. 105(3):640-657. doi: 10.1016/j.ajhg.2019.07.005. *Equal contribution
Gaston-Massuet, C., McCabe, M.J., Scagliotti, V., Young, R.M., Carreno, G., et al. (2016)
TCF7L1 is involved in hypothalamo-pituitary axis development.
PNAS. 2;113(5):E548-57. doi: 10.1073/pnas.1503346113.
Moro, E., Ozhan-Kizil G., Mongera, A., Beis, D., Wierzbicki, C., Young, R.M., et al. (2012)
In vivo Wnt signalling tracing through a transgenic biosensor fish reveals novel activity domains.
Dev. Biol. 366:327-40.
Andoniadou, C.L., Signore, M., Young, R.M., Gaston-Massuet, C., Fuchs, E., et al. (2011)
HESX1 and TCF3 mediated repression of Wntß-catenin targets is required for normal development of the anterior forebrain.
Development. 138:4931-4942.
Valdivia L.E.*, Young R.M.*±, Hawkins T.A., Stickney H.L., Cavodeassi F., et al. (2011)
Lef1-dependent Wnt/ß-catenin signalling drives the proliferative engine that maintains tissue homeostasis during lateral line development.
Development. 138(18):3931-41 * Equal contribution ±Co-corresponding author.
Ewan K, Pajak B, Stubbs M, Todd H, Barbeau O, Quevedo C, Botfield H, Young, R.M., et al. (2010)
A useful approach to identify novel small-molecule inhibitors of Wnt-dependent transcription.
Cancer Res.15;70(14):5963-73

Join Us/Contact
Join Us/Contact
Join Us/Contact Us
Interested in our research and/or working with us? Please contact: rodrigo.young@ucl.ac.uk
Our Lab is part of the
UCL Institute of Opthalmology
11-43 Bath Street
London
EC1V 9EL
Follow us on Bluesky
@younglab.bsKY.SOCIAL
Our Resources
Discover Zebrafish Research Lines
Shared academic catalogue of mutant and transgenic zebrafish lines held across UK research laboratories, supporting discovery, reuse and responsible collaboration.